Find measurable genetic biomarkers
Biomarker discovery
Find out about genetic biomarker discovery by NGS and microarrays
Genomic alterations form the basis of diseases and disorders. These alterations can be as small as single-nucleotide polymorphisms (SNPs), but also as extensive as thousands of bases of insertions or deletions (InDels) and copy number variants (CNVs) of whole genes.
Associated with disease and disorders are specific genetic biomarkers, which are DNA or RNA sequence, such as SNPs. These genetic biomarkers can be detected and could be used to draw conclusions on a person’s susceptibility to diseases and disorders (disease risk factors). They can also represent pharmacological targets.
The preferred methods for genetic biomarker discovery are next generation sequencing (NGS) and microarrays.
What options do you have for target identification
Applications for genetic biomarker discovery
Genome-wide genetic biomarker discovery with NGS
The genome-wide discovery of unknown genetic biomarkers is done by NGS-based whole genome sequencing, low pass whole genome sequencing and whole exome sequencing, as well as gene panels.
Whole genome sequencing (WGS) describes the sequencing of all DNA present in the genome (including mitochondrial DNA) and is performed on the Illumina NovaSeq 6000 platform with a 30x coverage. The sequencing results are then compared to a reference sequence to identify alteration between samples and enable biomarker discovery.

Image: Depiction of a single-nucleotide polymorphism in humans (image source: https://www.genome.gov/dna-day/15-ways/human-genomic-variation, created by: National Human Genome Research Institute).
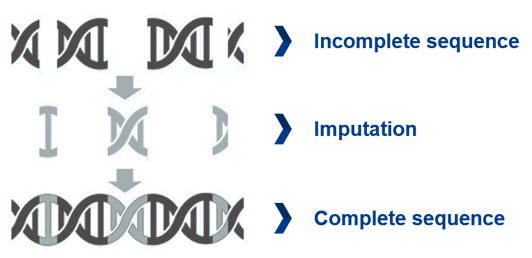
Image: Depiction of the low pass whole genome sequencing principle.
Low pass whole genome sequencing (LP-WGS) is a cost-efficient alternative to WGS for biomarker discovery. It is performed on the NovaSeq 6000 platform with a 10x coverage. Due to the lower coverage, some genomic regions will be incomplete. However, the missing genomic region can be filled by imputation algorithm. The faster sample throughput and lower costs make LP-WGS a great application to screen large cohorts with thousands of patients for instance.
Whole exome sequencing (WES) is widely utilised for biomarker discovery. As the exome represents about 1% of the whole human genome but contains approximately 85% of disease-related mutations, WES can be used to discover novel mutations and genetic biomarkers for complex diseases. Due to the small proportion of the exome compared to the genome, WES has a 100x average on target coverage.
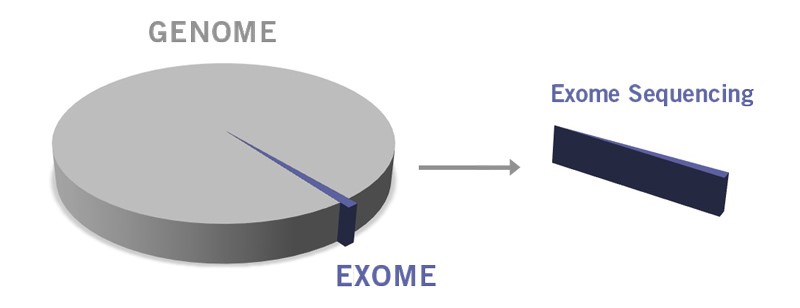
Image: Share of the exome compared to the whole genome.

Transcriptomics for genetic biomarker discovery
Genetic biomarkers can also be discovered on the RNA level. With a whole transcriptome analysis, researchers can gain access to all expressed RNAs, including miRNAs that have been proposed as biomarkers for different cancers and myocardial infarction for instance, and splicing events. Transcriptome analysis can be performed even if only small amounts of input RNA is available (down to 0.15 ng/µl).
By utilising RNA enrichment, transcriptome sequencing can also be directed to specific parts of the transcriptome for a more focused RNA biomarker discovery.
Genetic biomarker discovery with gene panels
Gene panels commonly describe the simultaneous NGS-based analysis of genes or regions (from a few to thousands) that are associated with specific diseases and disorders. Capturing of specific genes and regions is done by hybridisation to target-specific probes.
Target enrichment panels are used to analyse any genomic region, from the exome down to single genes, to detect SNPs and InDels, fusion genes and copy number variations (CNVs). Target enrichment panels (using hybridisation enrichment) are regarded by many as the best-suited method for genetic biomarker discovery and outcompete enrichment by amplicons. An outstanding example for target enrichment panels is the INVIEW Oncopanel that allows for the analysis of 591 key cancer-specific genes.
Customised enrichment panels are based on an enrichment according to the customer’s requirements for biomarker discovery. These customised panels enable a focused analysis of any genomic region of interest and specific lists of genes. They give researchers the highest amount of flexibility, including coverage and sensitivity to certain genetic biomarkers.
The design of target enrichment panels is done and validated by the Eurofins Genomics research and development (R&D) team.
Genetic biomarker discovery with microarrays
We offer genetic biomarker discovery with genome-wide genotyping microarrays that are built on whole-genome reference data from the “1000 Genomes Project”, a catalogue of human genetic variations (Illumina Global Screening Array v3.0 and Thermo Fisher Precision Medicine Research Array). These microarrays target genetic variants that are relevant for clinical research, including disease and trait associated markers.
Microarrays can also be used for the expression analysis of the coding transcriptome in high sample throughput. The utilised Clariom GO Screen arrays can detect around 20,000 genes without overlap for pathway analysis for instance. Significantly, these gene expression microarrays utilise cells to signal technology, where cell lysates are sufficient as samples. This gives researchers additional flexibility for their biomarker discovery projects.
Epigenetic biomarker discovery
The discovery of epigenetic biomarkers is done by whole genome bisulfite sequencing (WGBS), low pass whole genome bisulfite sequencing and microarrays. Here, cytosine methylation markers are commonly discovered by reduced representation bisulphite sequencing (RRBS) with an enrichment of CpG islands, target enrichment panels that focus on differentially methylated regions, and microarrays for the quantification of 850,000 methylation sites with extensive coverage of CpG islands, genes, and enhancers (EPIC array).
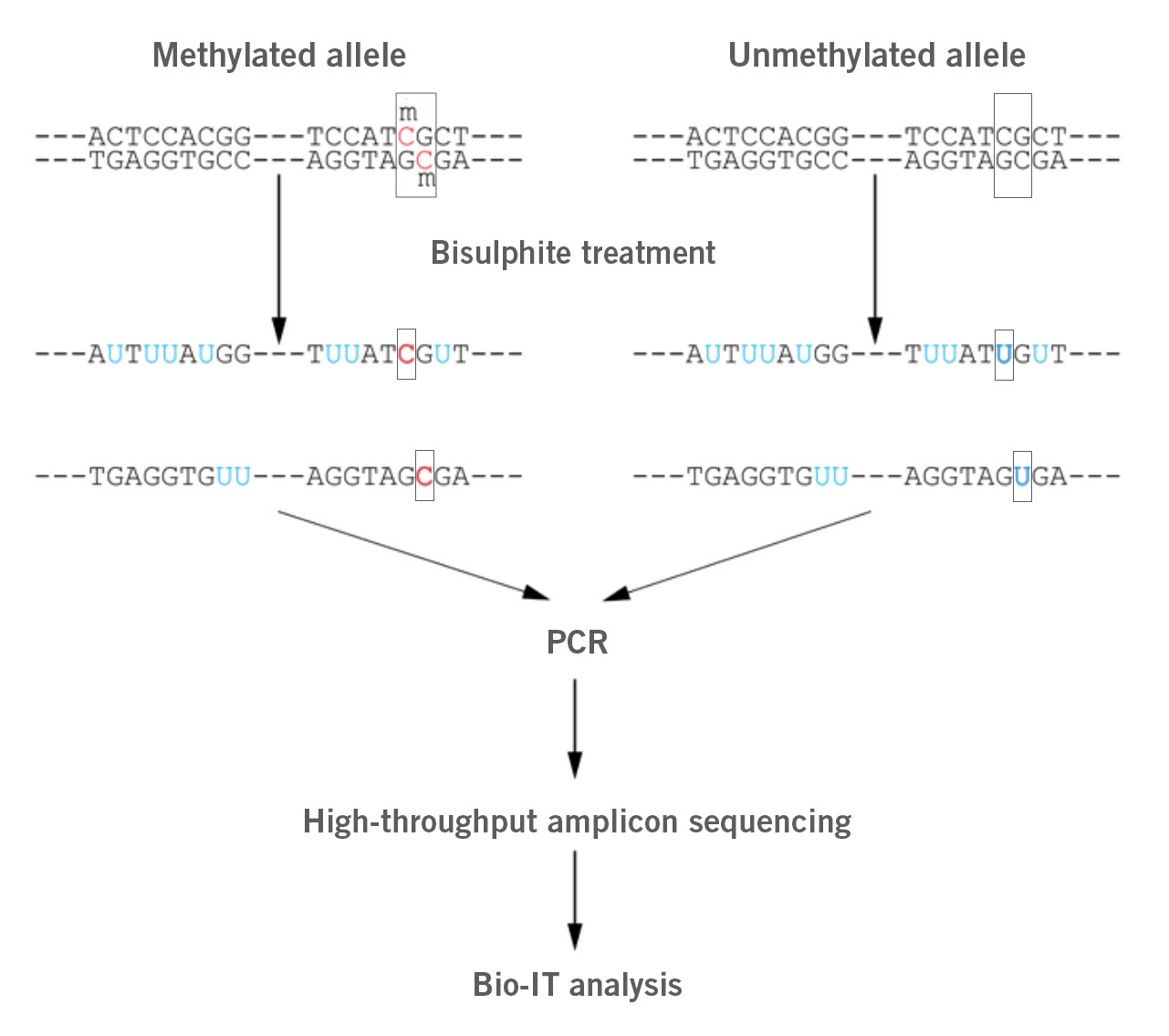
Image: Depiction of bisulphite sequencing process. Unmethylated cytosines are converted into uracil by bisulphite. Methylated cytosines are not converted. During the following PCR, all uracils are converted to thymidines. Subsequent amplicon sequencing with high sequencing coverage and mapping of the results to a reference genome reveal methylated loci.
After biomarkers have been discovered, they need to be validated in terms of accuracy, sensitivity and specificity for their intended application.
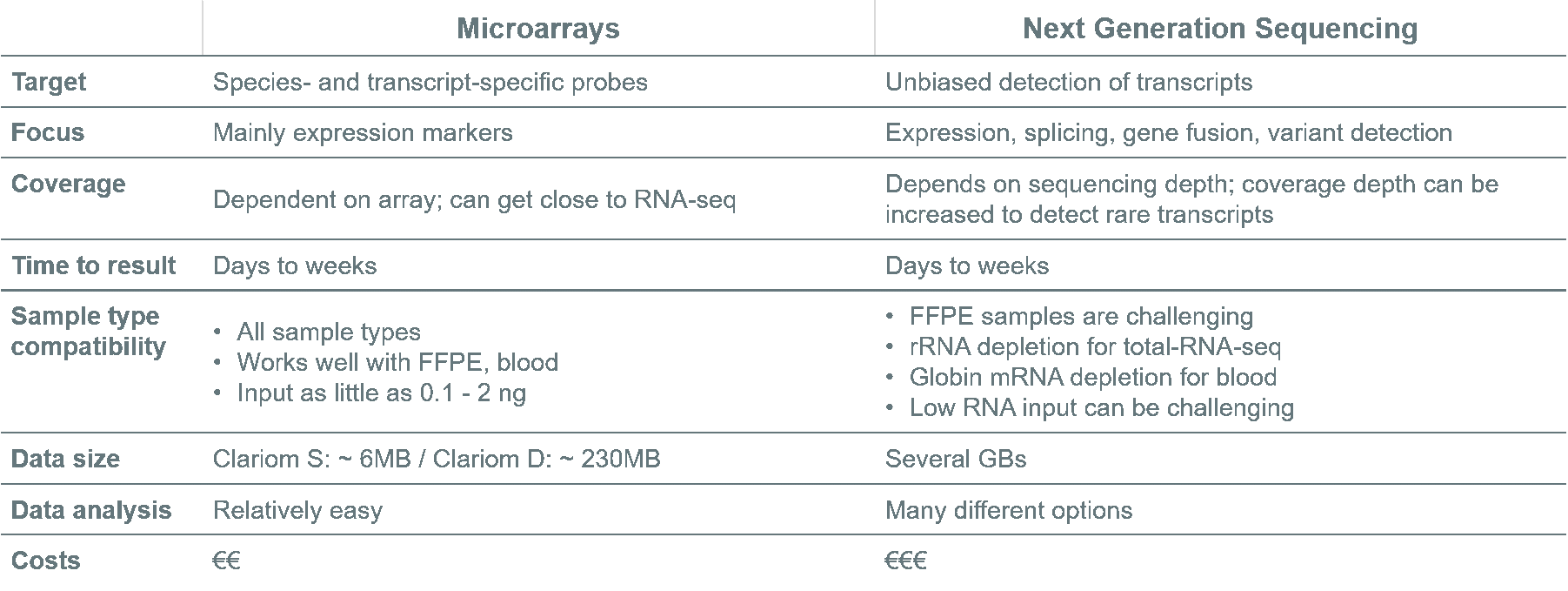
Genetic biomarker discovery – NGS or microarray
Researchers often ask the question if NGS or microarrays would suit their biomarker discovery projects best. The table below compares both methods and provides information to help researchers make decision.
Are you still not sure what method to choose?
Contact our pharmacogenomics expert for a free consultation now.
