Validate genetic biomarkers
Biomarker validation
Find out about genetic biomarker validation by microarrays and NGS
Diseases and disorders are mostly caused by genomic alterations that can range from single-nucleotide polymorphisms (SNPs) and insertions or deletions (InDels) to copy number variants (CNVs) of entire genes. These alterations can be identified, detected and used for disease susceptibility assessment.
In order to validate the discovered genetic biomarkers, next generation sequencing (NGS), microarrays, qPCR / dPCR and Sanger sequencing are used as preferred methods.
How to find the needle in the haystack
Applications for genetic biomarker validation
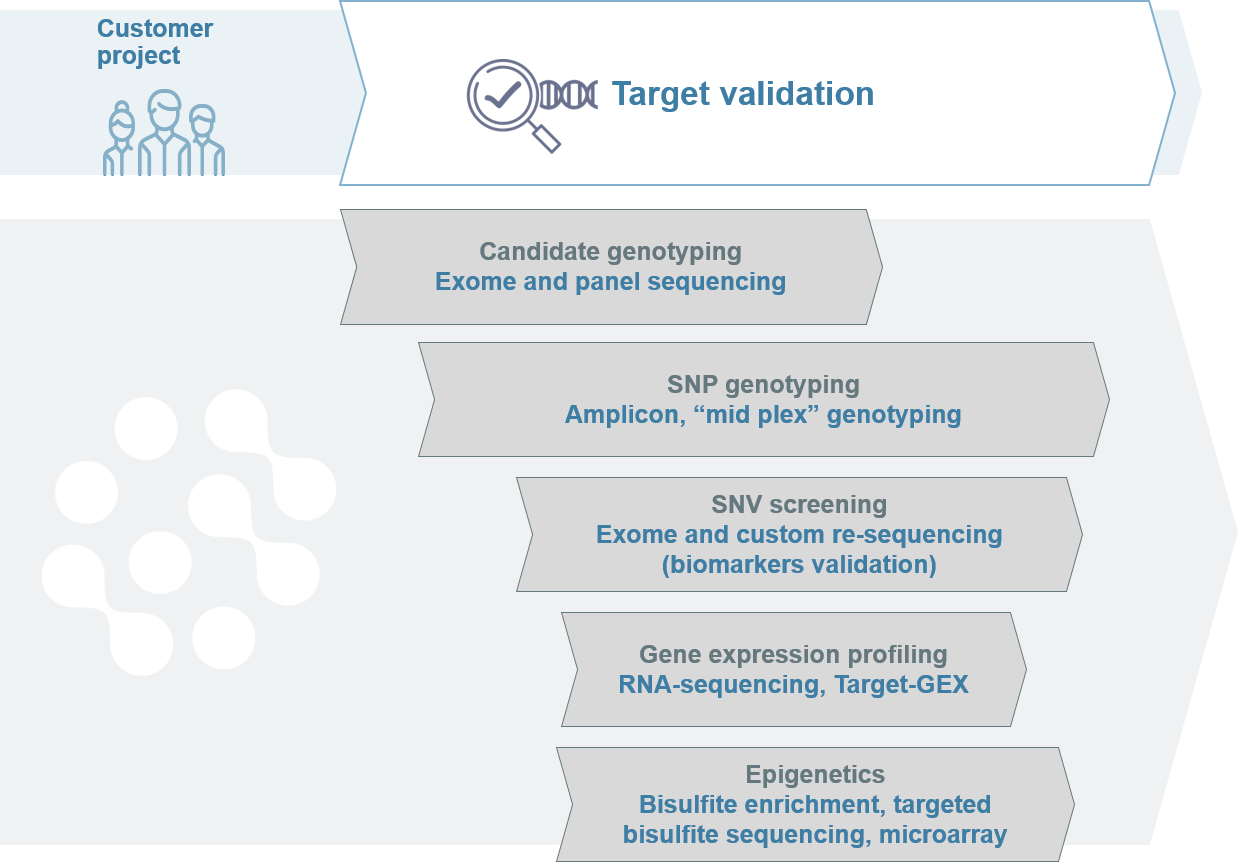
After a range of biomarkers have been identified, they need to be validated for accuracy, sensitivity and specificity in the context of their application.
During the discovery and validation stages, the number of applicable biomarkers decreases. Biomarkers must be able to differentiate tissues, such as diseased and healthy tissues, and predict the clinically relevant outcome in large cohorts.
Midplex genotyping
Genotyping of 10 to 1,000 markers with NGS-based midplex assays according to customer requirements.
The midplex assay is designed and validation by our biomarker experts using a subset of samples. In the lab, DNA is extracted from the samples in our highly automated extraction pipeline and prepared for the enrichment of the set of SNPs selected by the customer. High-throughput sequencing is performed on Illumina NovaSeq 6000 platforms and the data is analysed according to quantitative allele ratios.


Flexibility and customisation are important factors for any project. Midplex genotyping is no exception. We offer midplex assays for customised sets of SNPs (10 – 1,000), flexible addition of further SNPs at a later stage (when certain SNPs are found to be missing) and samples at any time. BioIT analyse can be done for either one SNP or several SNPs per target region.
High accuracy and robustness of our midplex assays are illustrated by a success rate of more than 97% for SNPs (on average) and a sample call rate of more than 98%.
The assays are very robust in terms of sample input, as only low DNA amounts, 20 ng, are needed (20 ng). Even crude lysates are usable for our midplex assays, as we are able to extract DNA from blood, tissue and cells.

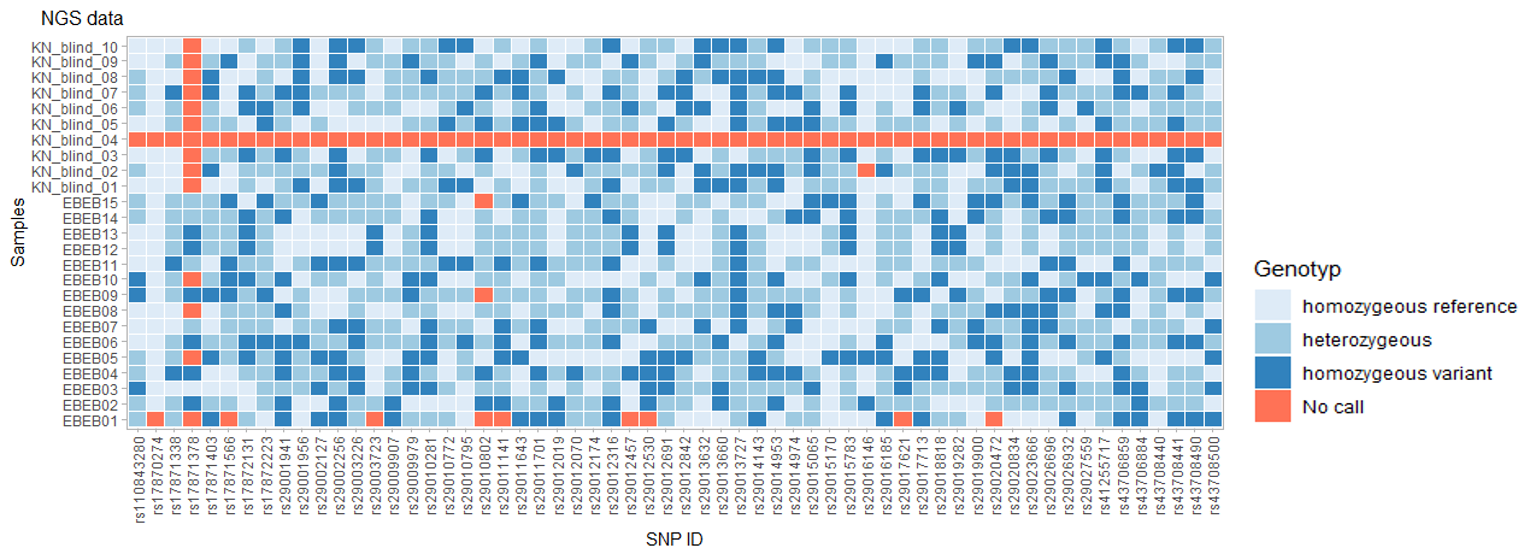
Midplex assay results
This example of a midplex assay design result with 60 SNPs shows great success with a call rate of 98.3% (99.2% with exclusion of one poorly performing SNP – rs17871338) and only one of 25 samples failed (KN_blind_04).
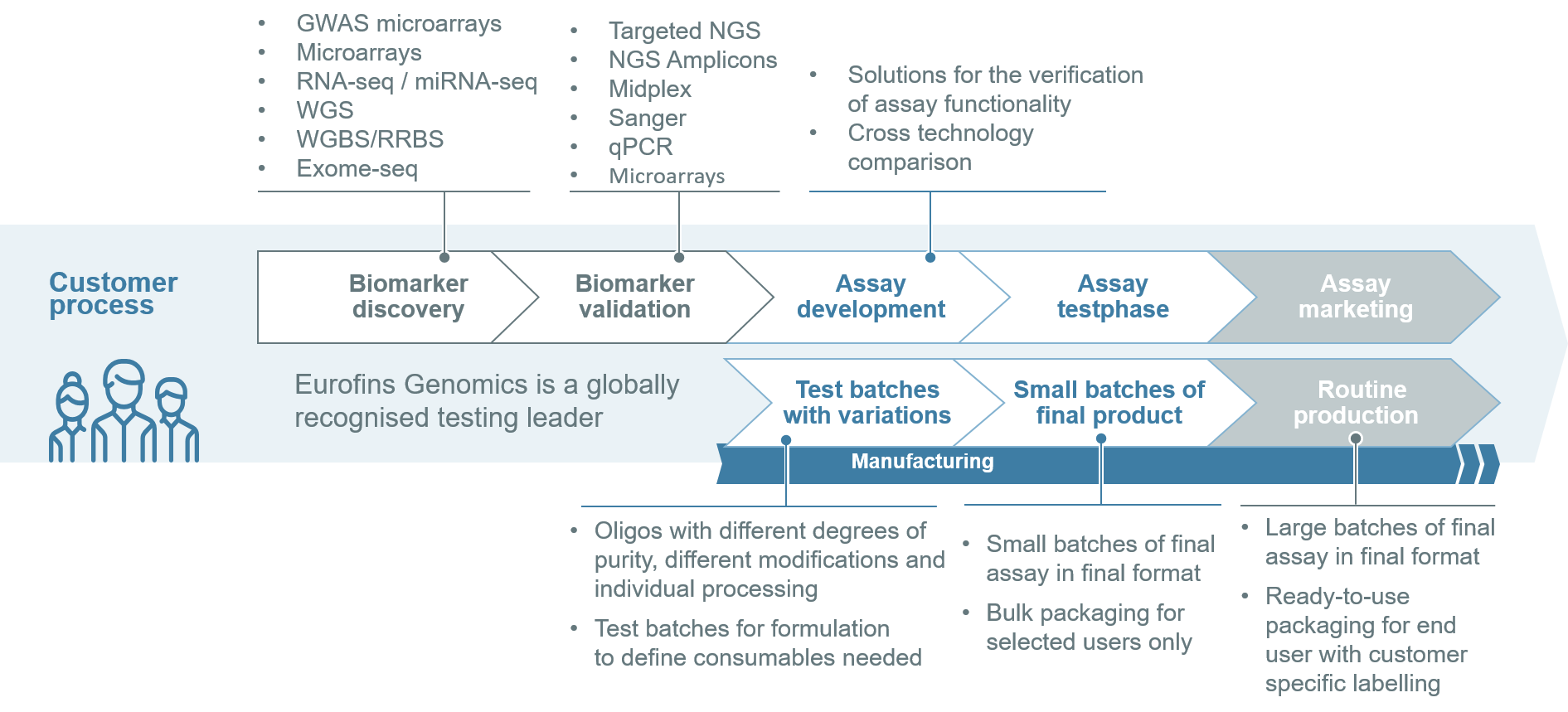
Genomics integrated approach
We cover the entire value chain of diagnostic test development.